GWENA: gene co-expression networks analysis and extended modules characterization in a single Bioconductor package, BMC Bioinformatics
Por um escritor misterioso
Last updated 31 março 2025

PDF] Gene co-expression analysis for functional classification and gene–disease predictions
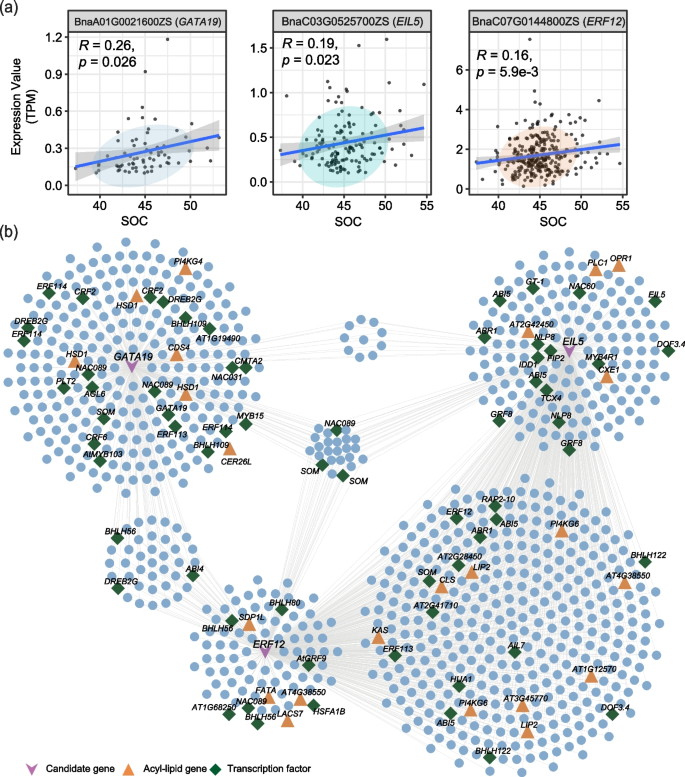
Interaction between phenylpropane metabolism and oil accumulation in the developing seed of Brassica napus revealed by high temporal-resolution transcriptomes, BMC Biology
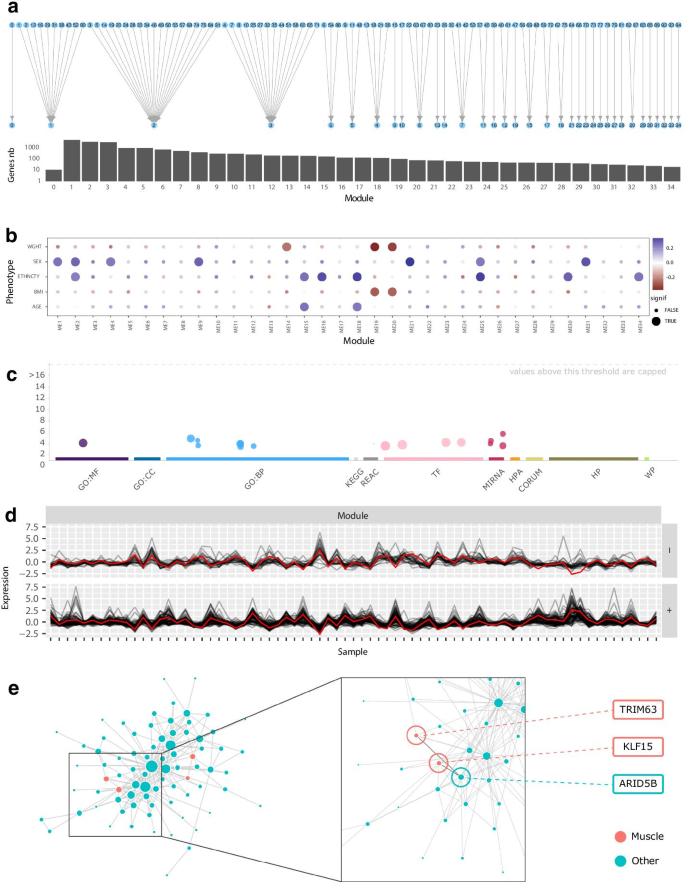
Single-Cell Co-expression Analysis Reveals Distinct Functional Modules, Co-regulation Mechanisms and Clinical Outcomes
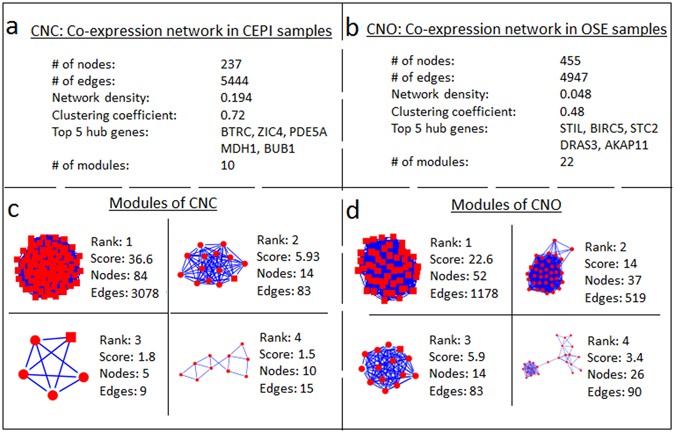
Differential co-expression analysis reveals a novel prognostic gene module in ovarian cancer
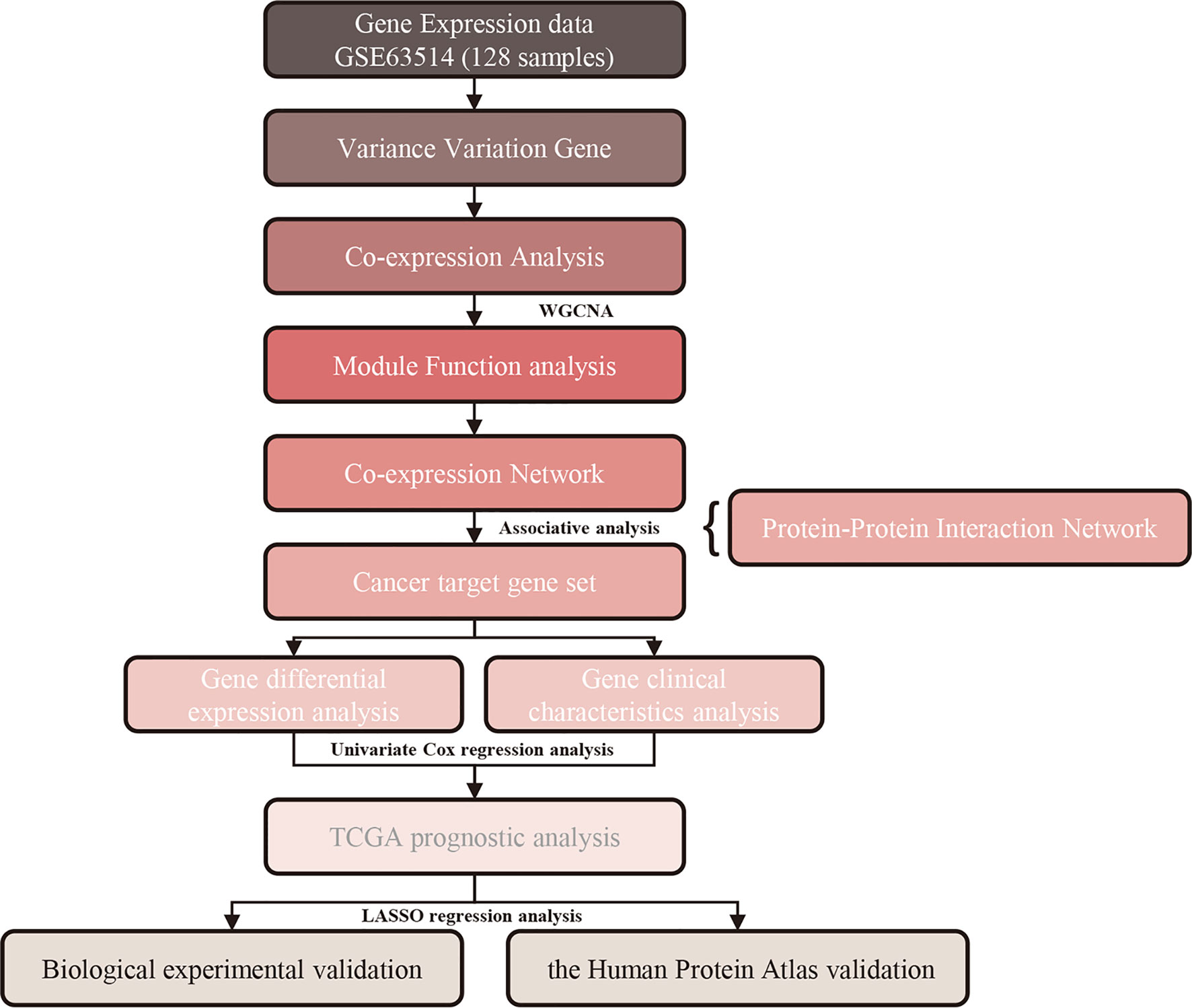
Frontiers Construction of Gene Modules and Analysis of Prognostic Biomarkers for Cervical Cancer by Weighted Gene Co-Expression Network Analysis

Bayesian Optimized sample-specific Networks Obtained By Omics data (BONOBO)

Topological properties of the Arabidopsis gene co-expresion network

Recent advances in the genetics underlying wheat grain protein content and grain protein deviation in hexaploid wheat - Paina - 2023 - Plant Biology - Wiley Online Library
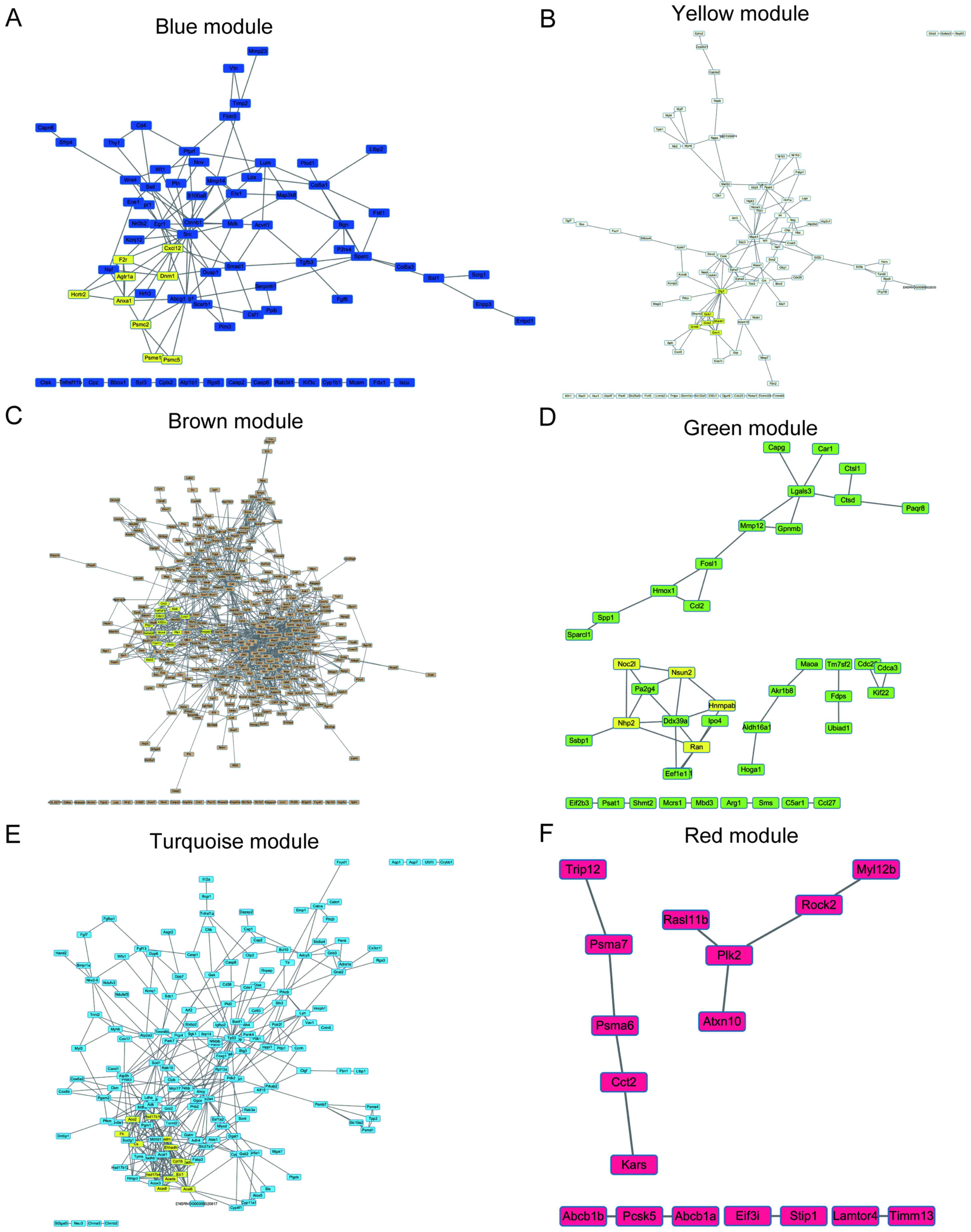
Weighted gene co‑expression network analysis in identification of key genes and networks for ischemic‑reperfusion remodeling myocardium

Clustering using WGCNA - Bioinformatics Team (BioITeam) at the University of Texas - UT Austin Wikis
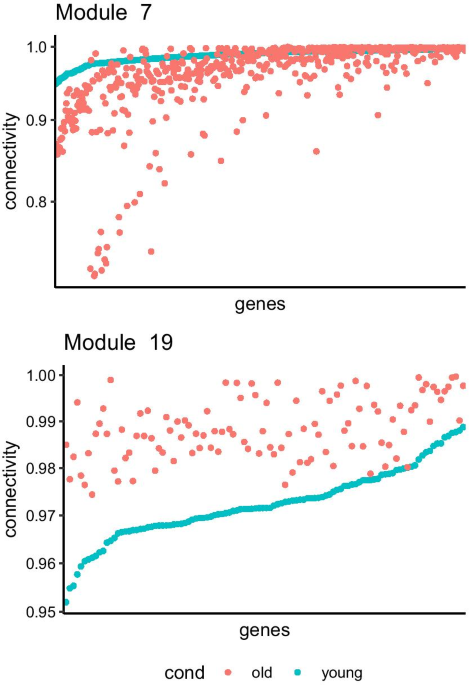
GWENA: gene co-expression networks analysis and extended modules characterization in a single Bioconductor package, BMC Bioinformatics
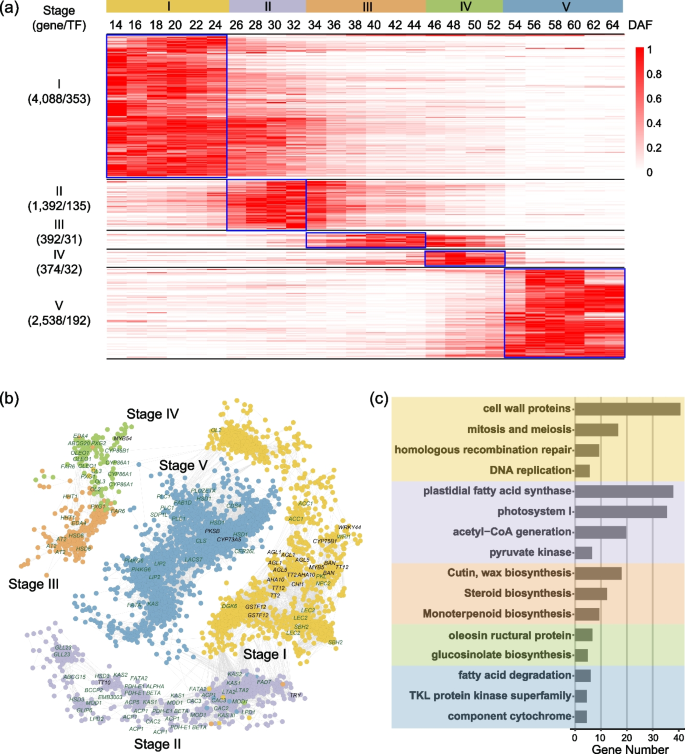
Interaction between phenylpropane metabolism and oil accumulation in the developing seed of Brassica napus revealed by high temporal-resolution transcriptomes, BMC Biology
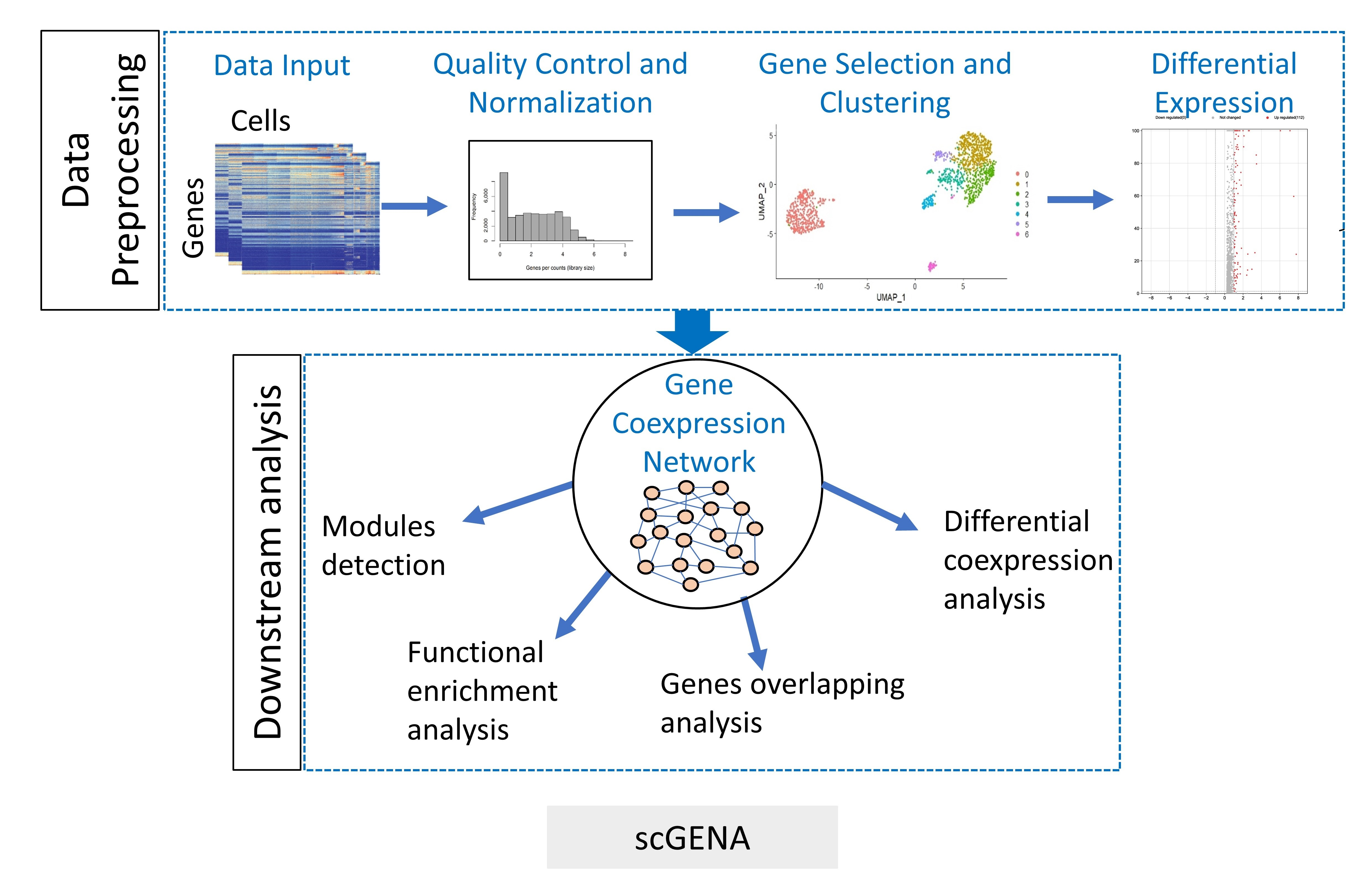
Bioengineering, Free Full-Text

PDF] Gene co-expression analysis for functional classification and gene–disease predictions

PDF) GCEN: An Easy-to-Use Toolkit for Gene Co-Expression Network Analysis and lncRNAs Annotation
Recomendado para você
-
 The Commons : Drivers of Change and Opportunities for Africa by Agence Française de Développement - Issuu31 março 2025
The Commons : Drivers of Change and Opportunities for Africa by Agence Française de Développement - Issuu31 março 2025 -
 Double Diffusion, Shear Instabilities, and Heat Impacts of a Pacific Summer Water Intrusion in the Beaufort Sea in: Journal of Physical Oceanography Volume 52 Issue 2 (2022)31 março 2025
Double Diffusion, Shear Instabilities, and Heat Impacts of a Pacific Summer Water Intrusion in the Beaufort Sea in: Journal of Physical Oceanography Volume 52 Issue 2 (2022)31 março 2025 -
 Transcriptomic signatures across human tissues identify functional rare genetic variation31 março 2025
Transcriptomic signatures across human tissues identify functional rare genetic variation31 março 2025 -
Bartley, W W Unfathomed Knowledge, Unmeasured Wealth Portrait - Optimized OCR, PDF31 março 2025
-
 vocab.txt · Geotrend/distilbert-base-en-pl-cased at 8d3f09ab0371534b7b7fd7e5ffb6998049ddd79231 março 2025
vocab.txt · Geotrend/distilbert-base-en-pl-cased at 8d3f09ab0371534b7b7fd7e5ffb6998049ddd79231 março 2025 -
Enduser Companes, PDF, Chief Executive Officer31 março 2025
-
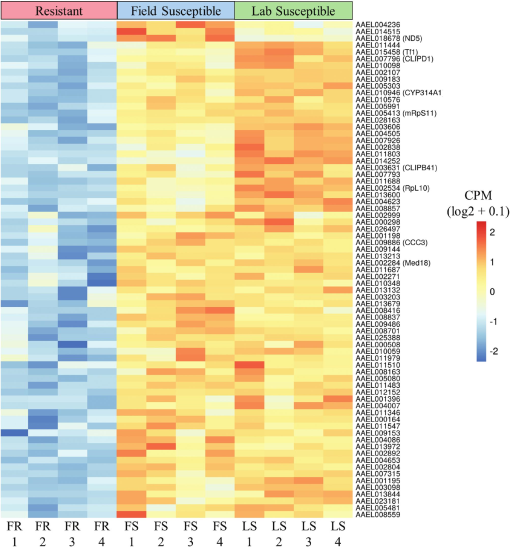 Expansive and Diverse Phenotypic Landscape of Field Aedes aegypti (Diptera: Culicidae) Larvae with Differential Susceptibility to Temephos: Beyond Metabolic Detoxification31 março 2025
Expansive and Diverse Phenotypic Landscape of Field Aedes aegypti (Diptera: Culicidae) Larvae with Differential Susceptibility to Temephos: Beyond Metabolic Detoxification31 março 2025 -
 PDF) Evolutionary Economics of Mental Time Travel?31 março 2025
PDF) Evolutionary Economics of Mental Time Travel?31 março 2025 -
 Springer Series on Touch and Haptic Systems31 março 2025
Springer Series on Touch and Haptic Systems31 março 2025 -
TimeFlow/settings/examples/5. Visualization NSF Grants (brief).time at master · FlowingMedia/TimeFlow · GitHub31 março 2025
você pode gostar
-
 Nintendo Direct Confirmed for Tomorrow - IGN31 março 2025
Nintendo Direct Confirmed for Tomorrow - IGN31 março 2025 -
 Vampire: The Masquerade: Bloodlines' fan patch is updated to include more content31 março 2025
Vampire: The Masquerade: Bloodlines' fan patch is updated to include more content31 março 2025 -
 Cooking Simulator VR - Welsh Cooking - SteamVR Game of 2021 : r/OculusLink31 março 2025
Cooking Simulator VR - Welsh Cooking - SteamVR Game of 2021 : r/OculusLink31 março 2025 -
 Cooking Simulator #2 (2021)31 março 2025
Cooking Simulator #2 (2021)31 março 2025 -
 Mochila Rosa bebé – peradoce31 março 2025
Mochila Rosa bebé – peradoce31 março 2025 -
 Preenchimento da Mandíbula - Dra. Malu Macedo - Agende sua avaliação31 março 2025
Preenchimento da Mandíbula - Dra. Malu Macedo - Agende sua avaliação31 março 2025 -
 video game websit by Shmuel Lipman31 março 2025
video game websit by Shmuel Lipman31 março 2025 -
 Tapete Pista Grande Cidade para Carrinhos Hot Wheels – Descrição Magazine31 março 2025
Tapete Pista Grande Cidade para Carrinhos Hot Wheels – Descrição Magazine31 março 2025 -
 Aesthetic wallpaper Pink retro wallpaper, Pink wallpaper iphone31 março 2025
Aesthetic wallpaper Pink retro wallpaper, Pink wallpaper iphone31 março 2025 -
 Keep It Live - Metacritic31 março 2025
Keep It Live - Metacritic31 março 2025