Modeling methyl-sensitive transcription factor motifs with an
Por um escritor misterioso
Last updated 06 janeiro 2025

Europe PMC is an archive of life sciences journal literature.
Possible scenarios by which DNA methylation could impact TF binding (A)

Abnormal methylation in the NDUFA13 gene promoter of breast cancer cells breaks the cooperative DNA recognition by transcription factors, QRB Discovery
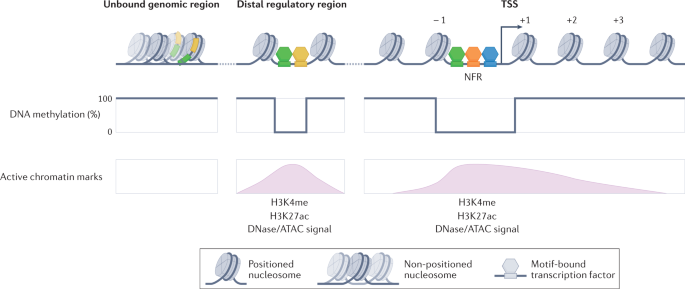
Generating specificity in genome regulation through transcription factor sensitivity to chromatin
Rare genetic variation at transcription factor binding sites modulates local DNA methylation profiles

Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet
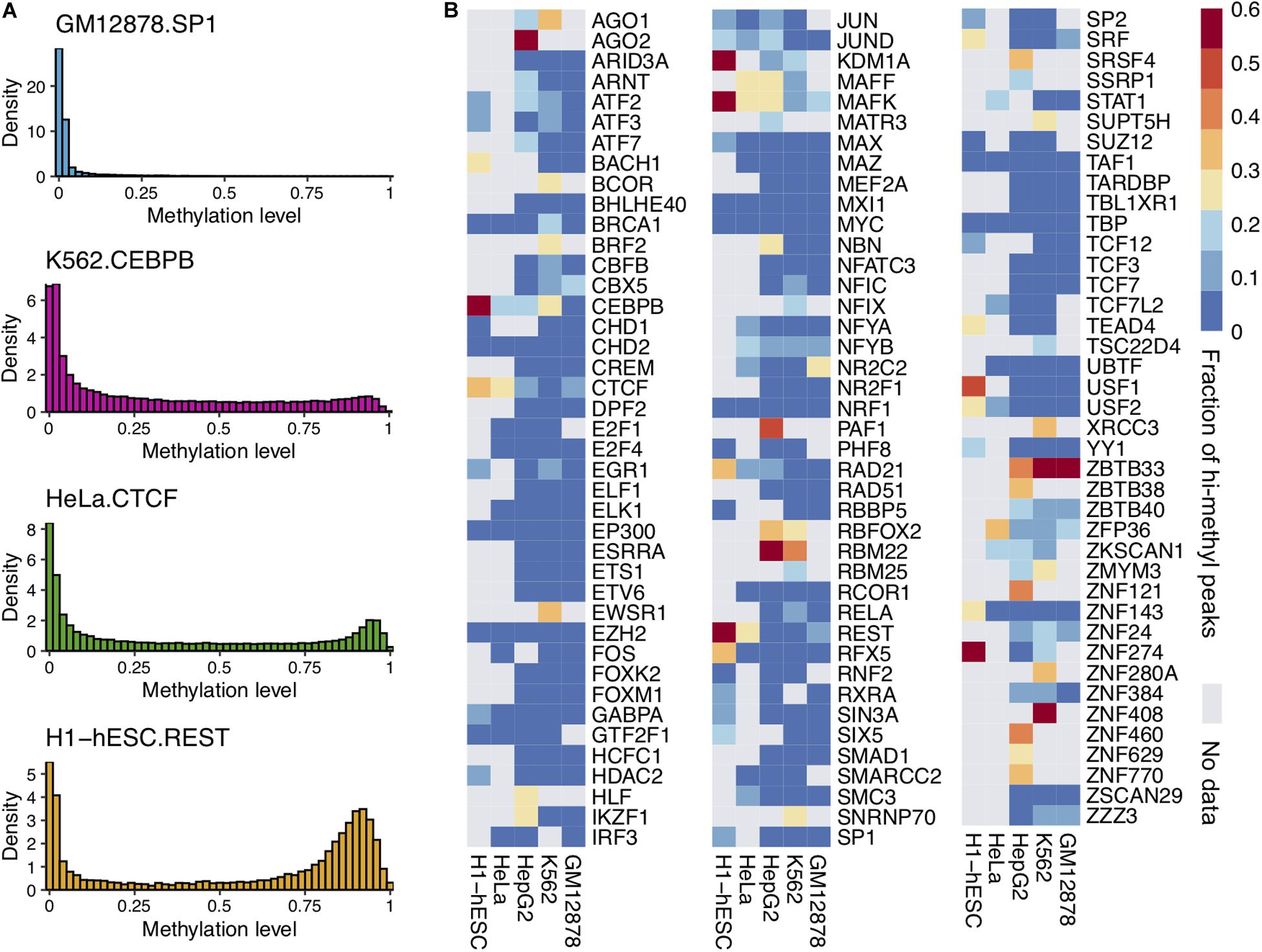
Frontiers Effects of DNA Methylation on TFs in Human Embryonic Stem Cells

Quantitative Analysis of the DNA Methylation Sensitivity of Transcription Factor Complexes - ScienceDirect

Physicochemical models of protein–DNA binding with standard and modified base pairs

A base-resolution panorama of the in vivo impact of cytosine methylation on transcription factor binding

Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet
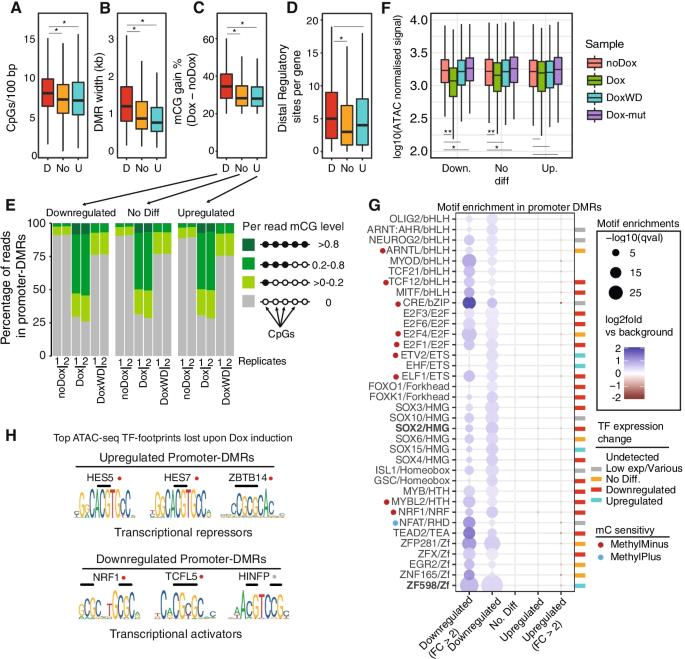
Large-scale manipulation of promoter DNA methylation reveals context-specific transcriptional responses and stability, Genome Biology
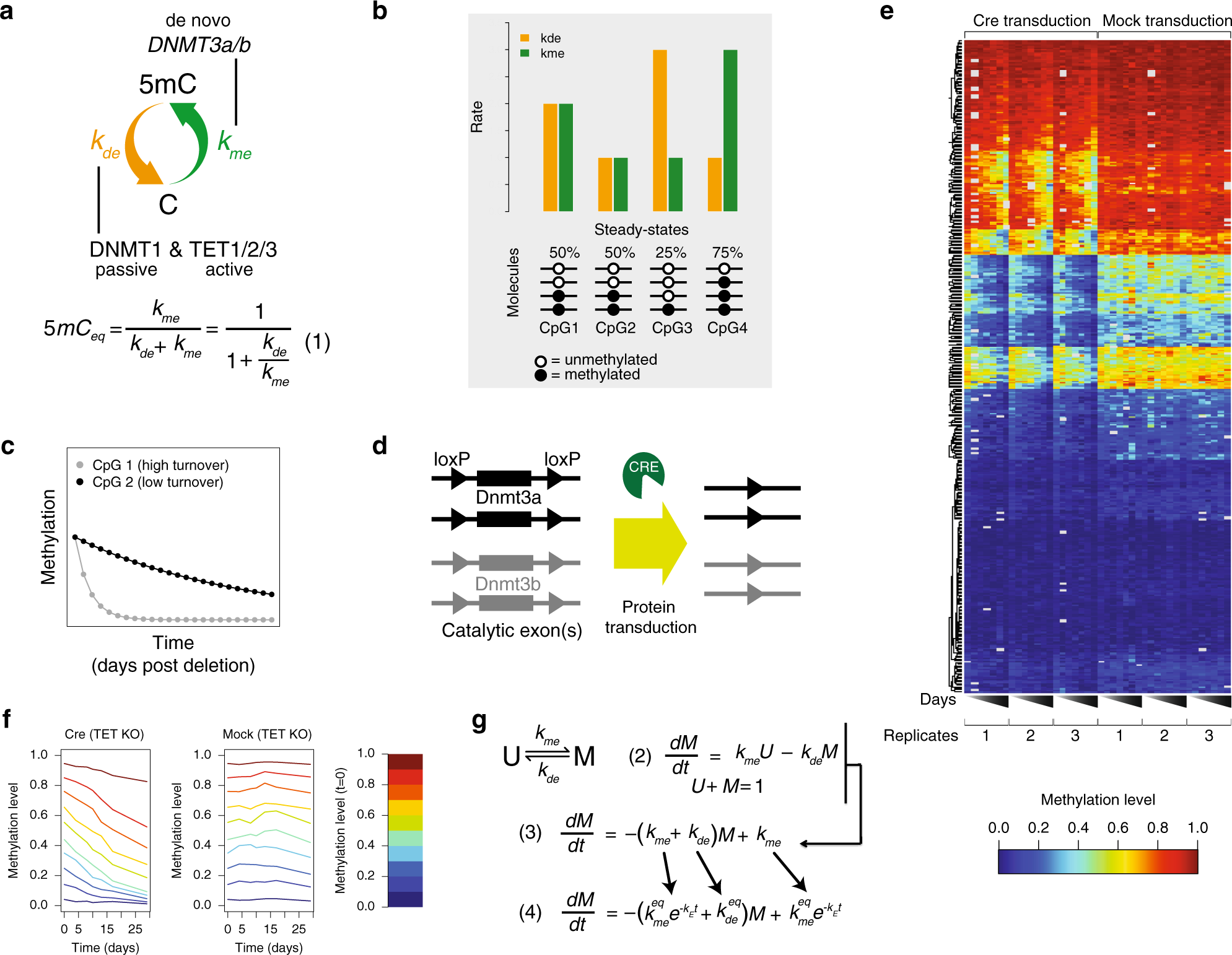
A genome-scale map of DNA methylation turnover identifies site-specific dependencies of DNMT and TET activity
Recomendado para você
-
 GTMAT Skull Smoking on The Sunset Beach Bathroom Rugs,Soft Absorbent Bath Mat,Machine Washable Dry Bath Mats for Indoor Living Room, Tub and Shower06 janeiro 2025
GTMAT Skull Smoking on The Sunset Beach Bathroom Rugs,Soft Absorbent Bath Mat,Machine Washable Dry Bath Mats for Indoor Living Room, Tub and Shower06 janeiro 2025 -
 Curso de Análise Combinatória e Probabilidade – GTMAT06 janeiro 2025
Curso de Análise Combinatória e Probabilidade – GTMAT06 janeiro 2025 -
50 SQFT GTMAT PRO 50MIL ROLL (18 X 33.3') AUTOMOTIVE AUDIO SOUND06 janeiro 2025
-
 GTS Style Vespa Floor Mat For GT GTS GTV Super 300 HPE 602734M06 janeiro 2025
GTS Style Vespa Floor Mat For GT GTS GTV Super 300 HPE 602734M06 janeiro 2025 -
 Divulgação da equipe – edição 2022 – GTMAT06 janeiro 2025
Divulgação da equipe – edição 2022 – GTMAT06 janeiro 2025 -
How to generate gt.mat file for new dataset? · Issue #220 · ankush06 janeiro 2025
-
![Image 7 of New-York tribune (New York [N.Y.]), August 3, 1908](http://tile.loc.gov/image-services/iiif/service:ndnp:dlc:batch_dlc_brazil_ver01:data:sn83030214:00175039387:1908080301:0078/full/pct:12.5/0/default.jpg) Image 7 of New-York tribune (New York [N.Y.]), August 3, 190806 janeiro 2025
Image 7 of New-York tribune (New York [N.Y.]), August 3, 190806 janeiro 2025 -
 My 2013 Complete Stereo Upgrade with costs06 janeiro 2025
My 2013 Complete Stereo Upgrade with costs06 janeiro 2025 -
 GTMAT Automotive Sound Dampener 50mil Pro Door Kit06 janeiro 2025
GTMAT Automotive Sound Dampener 50mil Pro Door Kit06 janeiro 2025 -
 Rockville Rockmat RM3 (2) 12x12 Butyl Rubber Sound Dampening/Deadening Car Kit06 janeiro 2025
Rockville Rockmat RM3 (2) 12x12 Butyl Rubber Sound Dampening/Deadening Car Kit06 janeiro 2025
você pode gostar
-
 Nintendo Accounts to 'Help Ease Transition' to the Switch Successor06 janeiro 2025
Nintendo Accounts to 'Help Ease Transition' to the Switch Successor06 janeiro 2025 -
Frases para o 4 ano - Recursos de ensino06 janeiro 2025
-
 007 Goldeneye Nintendo 64 GOTY Ed. CGC 9.0 A+ - goldencollectors06 janeiro 2025
007 Goldeneye Nintendo 64 GOTY Ed. CGC 9.0 A+ - goldencollectors06 janeiro 2025 -
 Tênis Masculino Cavalera Shark Branco/Laranja – 59110244 – Hang Store06 janeiro 2025
Tênis Masculino Cavalera Shark Branco/Laranja – 59110244 – Hang Store06 janeiro 2025 -
 EVIL DEAD RISE Official Trailer (2023) Horror Movie HD06 janeiro 2025
EVIL DEAD RISE Official Trailer (2023) Horror Movie HD06 janeiro 2025 -
/cdn.vox-cdn.com/uploads/chorus_asset/file/4117184/philspencer2.0.jpg) Read Microsoft Gaming CEO's email to staff about the Activision Blizzard acquisition - The Verge06 janeiro 2025
Read Microsoft Gaming CEO's email to staff about the Activision Blizzard acquisition - The Verge06 janeiro 2025 -
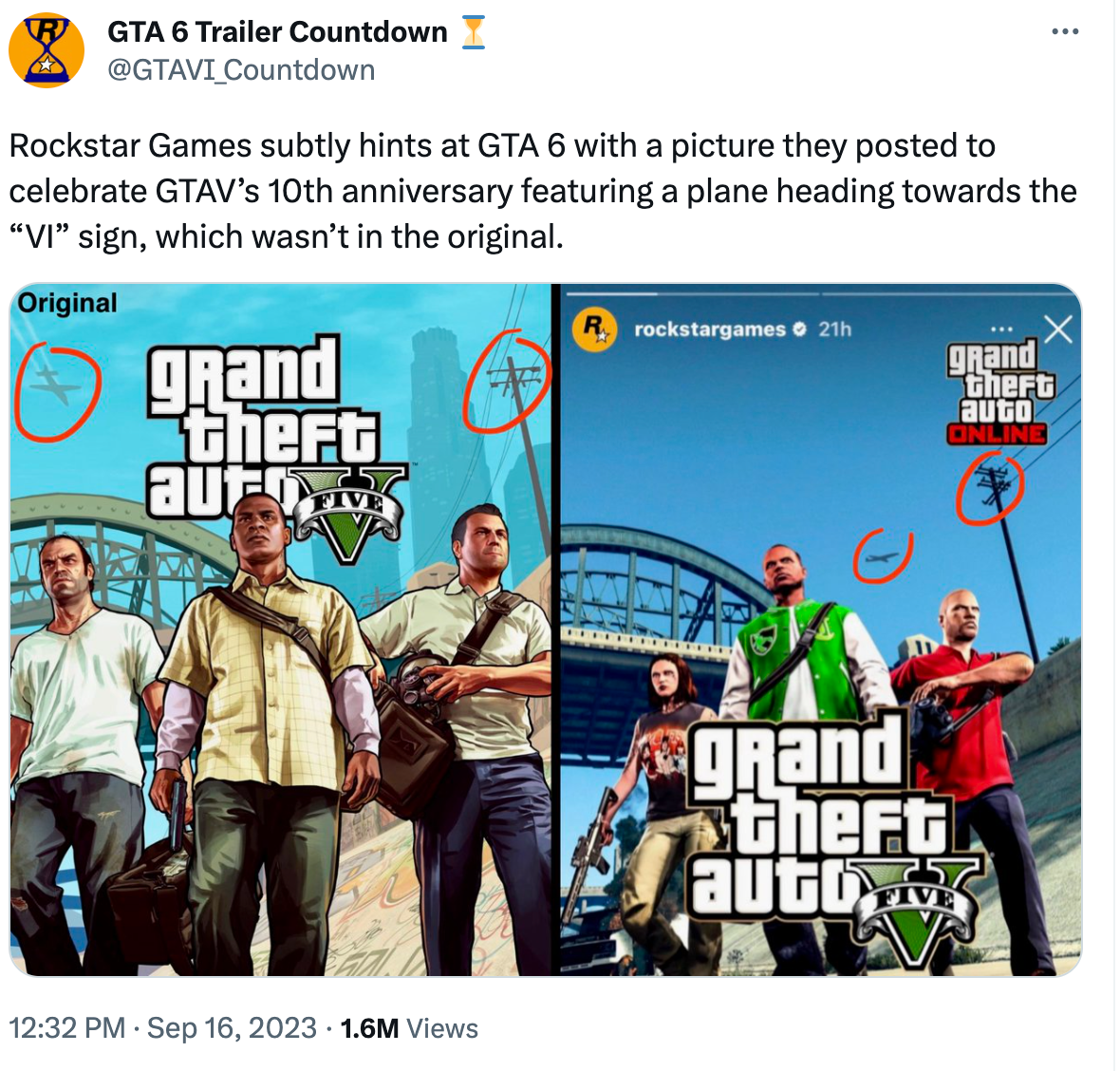 Fans think Rockstar Games hinted at GTA 6 with 10th anniversary photo06 janeiro 2025
Fans think Rockstar Games hinted at GTA 6 with 10th anniversary photo06 janeiro 2025 -
 Boneco Dragon Ball Z Goku Super Saiyajin Deus Action Figure06 janeiro 2025
Boneco Dragon Ball Z Goku Super Saiyajin Deus Action Figure06 janeiro 2025 -
 When are the Semifinals and Finals of the Worlds: Date, Time, Matches and Teams of the LoL Worlds - Meristation06 janeiro 2025
When are the Semifinals and Finals of the Worlds: Date, Time, Matches and Teams of the LoL Worlds - Meristation06 janeiro 2025 -
 Jakks Pacific Is Racing to Bring New Sonic Prime Toys to Fans - The06 janeiro 2025
Jakks Pacific Is Racing to Bring New Sonic Prime Toys to Fans - The06 janeiro 2025
