MVLs. (A and B) The structure (A) and molecular modeling (B) of
Por um escritor misterioso
Last updated 25 março 2025


MVLs. (A and B) The structure (A) and molecular modeling (B) of
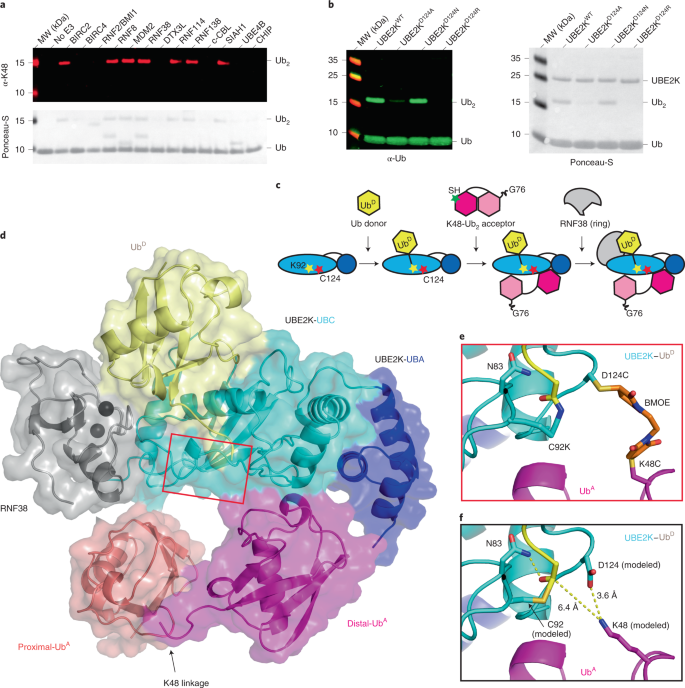
Structure of UBE2K–Ub/E3/polyUb reveals mechanisms of K48-linked Ub chain extension

Molecules May-1 2019 - Browse Articles
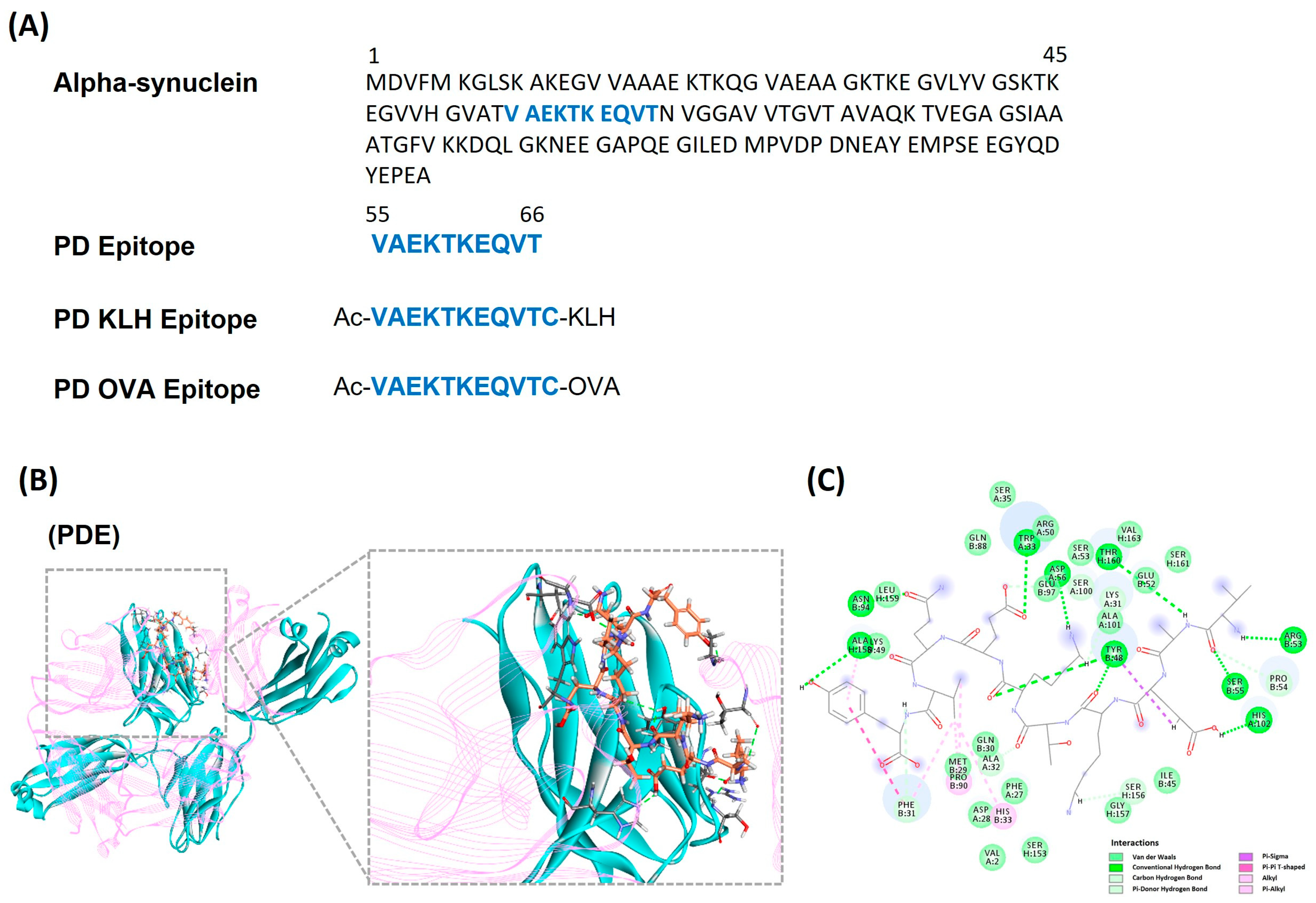
Vaccines, Free Full-Text
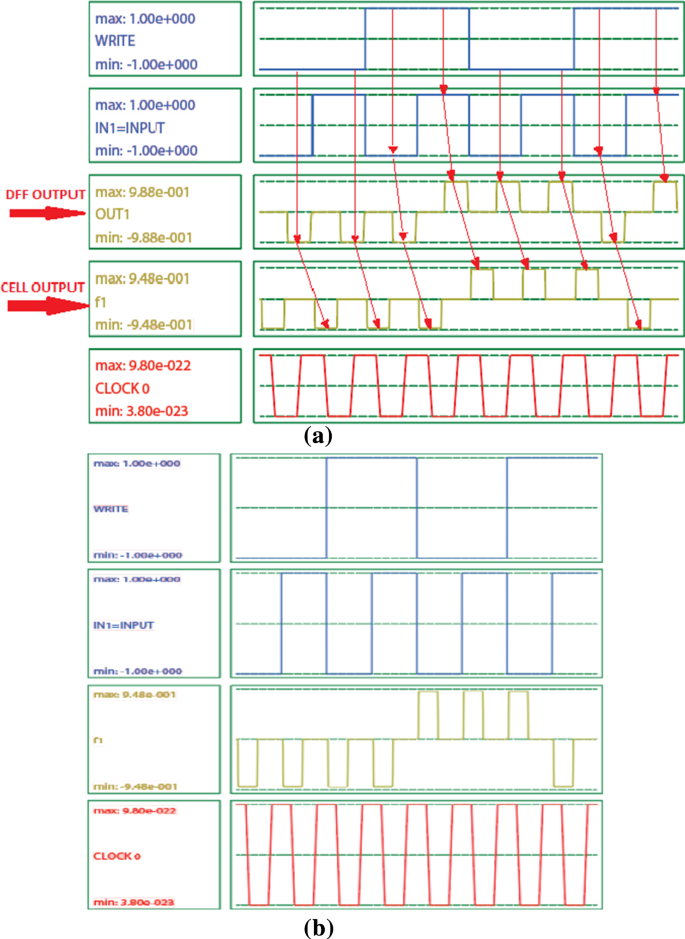
Realization of processing-in-memory using binary and ternary quantum-dot cellular automata

Solved e.com/drive/u/1/my-drive EXERCISES VİSUALIZING
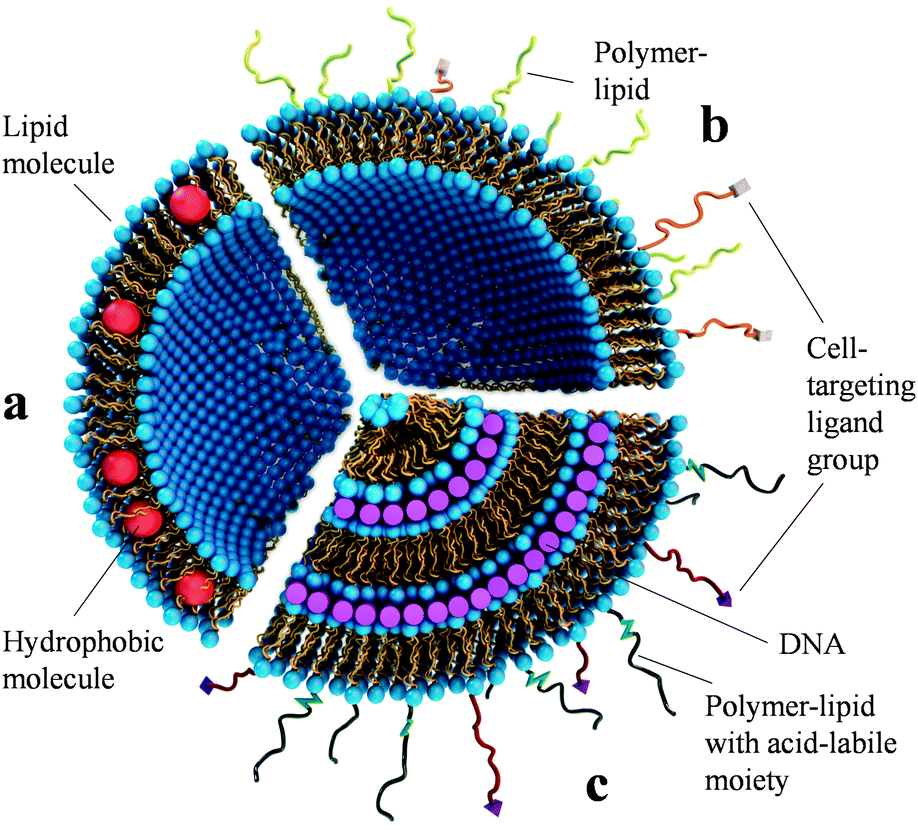
Cationic liposome–nucleic acid complexes for gene delivery and gene silencing - New Journal of Chemistry (RSC Publishing) DOI:10.1039/C4NJ01314J
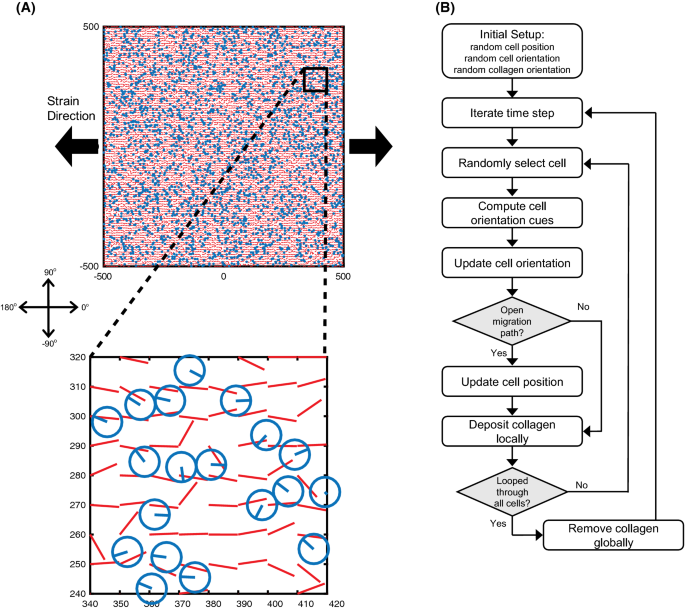
Potential strain-dependent mechanisms defining matrix alignment in healing tendons

Cationic liposome-nucleic acid complexes for gene delivery and silencing: pathways and mechanisms for plasmid DNA and siRNA. - Abstract - Europe PMC

Simulation of Self-Assembly of Cationic Lipids and DNA into Structured Complexes
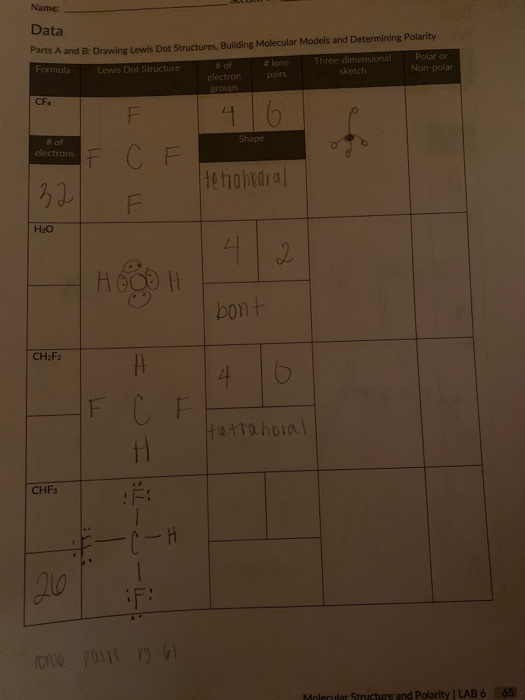
Solved Data Parts A and B: Drawing Lewis Dot Structures
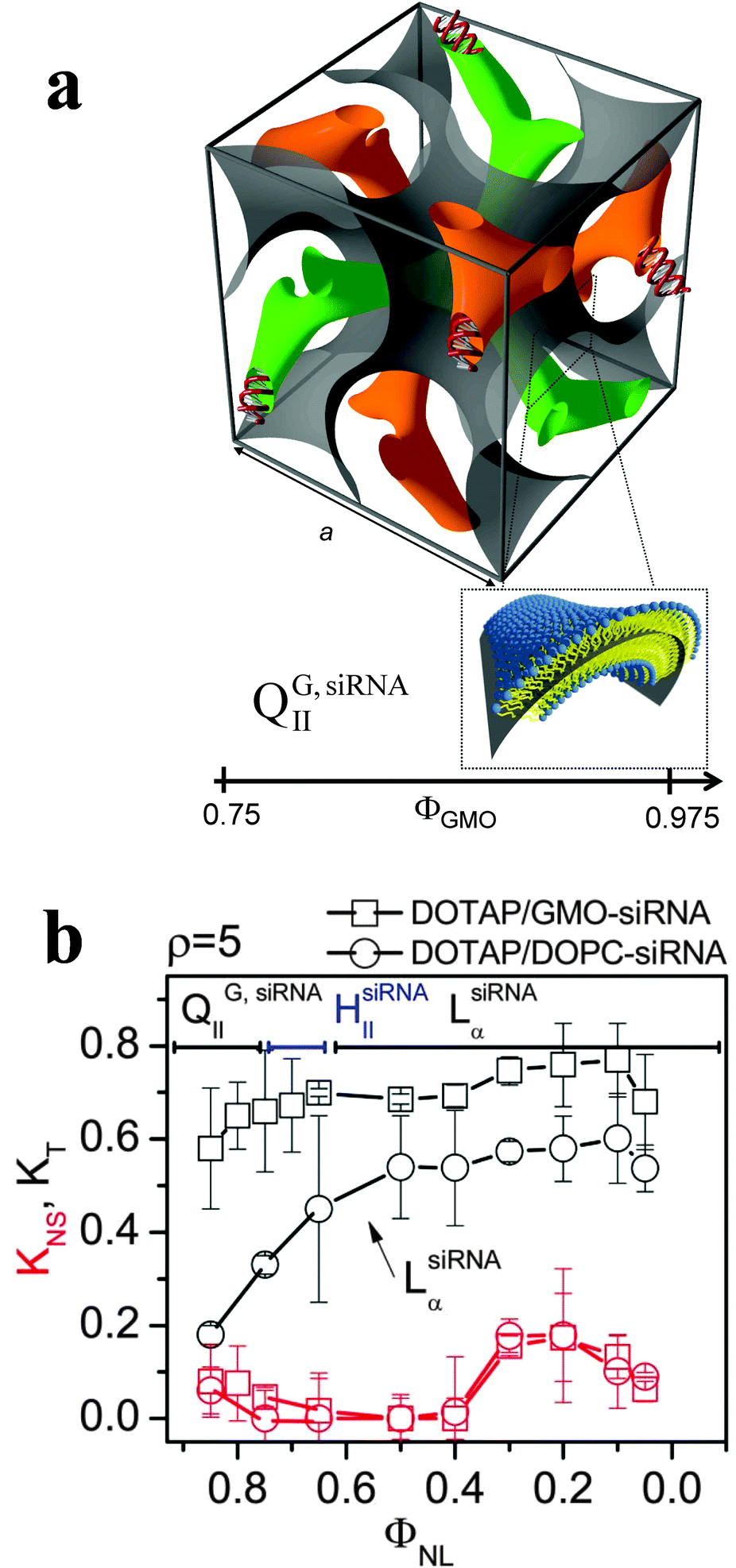
Cationic liposome–nucleic acid complexes for gene delivery and gene silencing - New Journal of Chemistry (RSC Publishing) DOI:10.1039/C4NJ01314J

A) In vitro release of Bev-MVLs (a) in three different mediums: normal
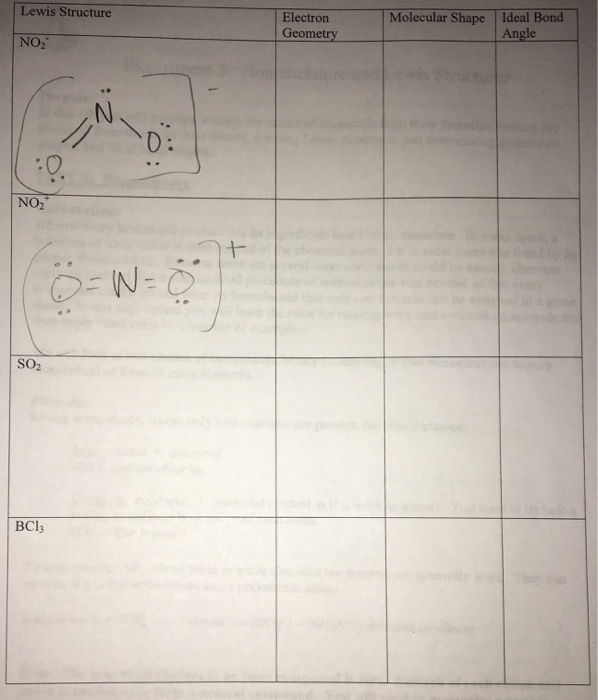
Solved PART B: Lewis Structures In the table provided, draw

The influence of the stereochemistry and C-end chemical modification of dermorphin derivatives on the peptide-phospholipid interactions - ScienceDirect
Recomendado para você
-
 Maryland Volunteer Lawyers Service (MVLS)25 março 2025
Maryland Volunteer Lawyers Service (MVLS)25 março 2025 -
Maryland Volunteer Lawyers Service (MVLS)25 março 2025
-
MVLS - Primary Incision, Various Artists25 março 2025
-
 Maryland Volunteer Lawyers Service Celebrate Pro Bono25 março 2025
Maryland Volunteer Lawyers Service Celebrate Pro Bono25 março 2025 -
Maryland Volunteer Lawyers Service25 março 2025
-
 MVLS AACPLL Blog: Law Library News25 março 2025
MVLS AACPLL Blog: Law Library News25 março 2025 -
 MVLS PGT College Induction - 18 September 201925 março 2025
MVLS PGT College Induction - 18 September 201925 março 2025 -
 Test Lomke Legal25 março 2025
Test Lomke Legal25 março 2025 -
 Marvelous Entertainment Wedding DJ - View 8 Reviews and 22 Pictures25 março 2025
Marvelous Entertainment Wedding DJ - View 8 Reviews and 22 Pictures25 março 2025 -
 BGL Sponsors MVLS 2023 Taste for Pro Bono - Brown Goldstein Levy25 março 2025
BGL Sponsors MVLS 2023 Taste for Pro Bono - Brown Goldstein Levy25 março 2025
você pode gostar
-
 Assistir Kaizoku Oujo Episodio 8 Online25 março 2025
Assistir Kaizoku Oujo Episodio 8 Online25 março 2025 -
 MIRROR - Japanese Tattoo Pin - Limited Edition Collaboration Monica Mo25 março 2025
MIRROR - Japanese Tattoo Pin - Limited Edition Collaboration Monica Mo25 março 2025 -
 37cm Pj Pug A Pillar Plush Poppy Playtime Caterpillar Peluche Huggy Wuggy Stuffed Animal Doll25 março 2025
37cm Pj Pug A Pillar Plush Poppy Playtime Caterpillar Peluche Huggy Wuggy Stuffed Animal Doll25 março 2025 -
Sonic Drift 2 Sega Game Gear For Sale25 março 2025
-
 Pin on game PlayStation25 março 2025
Pin on game PlayStation25 março 2025 -
 Family Feud Used PS2 Games For Sale Retro Game Store25 março 2025
Family Feud Used PS2 Games For Sale Retro Game Store25 março 2025 -
 Nintendo DSi XL Brown System - Discounted25 março 2025
Nintendo DSi XL Brown System - Discounted25 março 2025 -
 CapCut_jj kakashi25 março 2025
CapCut_jj kakashi25 março 2025 -
 Número do WhatsApp da Luluca (2023)25 março 2025
Número do WhatsApp da Luluca (2023)25 março 2025 -
 Caca Palavras - Nivel Facil-medio-dificil - Vol. 3 - 978857902787125 março 2025
Caca Palavras - Nivel Facil-medio-dificil - Vol. 3 - 978857902787125 março 2025



